Docker
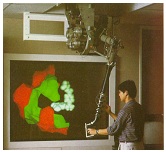 The Docker program was developed by
Ming Ouhyoung as part of his dissertation. This system (also
called GROPE) allows a chemist to interact (graphically and
haptically) with a real-time simulation of a drug molecule
interacting with its receptor site in a protein.
The Docker program was developed by
Ming Ouhyoung as part of his dissertation. This system (also
called GROPE) allows a chemist to interact (graphically and
haptically) with a real-time simulation of a drug molecule
interacting with its receptor site in a protein.
This system is being used and extended at NCSA as the IDock
Molecular Docking project.
Trailblazer
Over the past twenty-five years, GRIP has built more than a
dozen computer graphics systems for exploring new ideas in
molecular graphics, that is, to blaze new trails. Many of these
systems have incorporated distinct software components that have
similar or even identical function, such as the display of
molecular structure and calculation of interatomic forces. The
Trailblazer project is building an infrastructure of
object-oriented data structures and routines with which to build
exploratory systems more quickly and more surely. This
infrastructure needs refinement and testing within the context of
real application programs. The first will be a Docker tool for
exploring how drugs bind in and act on proteins.
¡@
(Note: The above is from the Web page of UNC-Chapel Hill,
Dept. of CS,
http://www.cs.unc.edu/Research/nano/cismm)
 The Docker program was developed by
Ming Ouhyoung as part of his dissertation. This system (also
called GROPE) allows a chemist to interact (graphically and
haptically) with a real-time simulation of a drug molecule
interacting with its receptor site in a protein.
The Docker program was developed by
Ming Ouhyoung as part of his dissertation. This system (also
called GROPE) allows a chemist to interact (graphically and
haptically) with a real-time simulation of a drug molecule
interacting with its receptor site in a protein.